- Review
- Open access
- Published:
It is time for top-down venomics
Journal of Venomous Animals and Toxins including Tropical Diseases volume 23, Article number: 44 (2017)
Abstract
The protein composition of animal venoms is usually determined by peptide-centric proteomics approaches (bottom-up proteomics). However, this technique cannot, in most cases, distinguish among toxin proteoforms, herein called toxiforms, because of the protein inference problem. Top-down proteomics (TDP) analyzes intact proteins without digestion and provides high quality data to identify and characterize toxiforms. Denaturing top-down proteomics is the most disseminated subarea of TDP, which performs qualitative and quantitative analyzes of proteoforms up to ~30 kDa in high-throughput and automated fashion. On the other hand, native top-down proteomics provides access to information on large proteins (> 50 kDA) and protein interactions preserving non-covalent bonds and physiological complex stoichiometry. The use of native and denaturing top-down venomics introduced novel and useful techniques to toxinology, allowing an unprecedented characterization of venom proteins and protein complexes at the toxiform level. The collected data contribute to a deep understanding of venom natural history, open new possibilities to study the toxin evolution, and help in the development of better biotherapeutics.
Background
Venom is a complex mixture of proteins and other chemical compounds used to paralyze or kill prey and to subjugate predators [1]. Its composition generally presents a range of a few to dozens of toxin families playing the most diverse pharmacological functions [2]. Animal toxins encoded by several multiloci gene families result in a large number of expressed protein forms that can differ greatly among individuals even from the same species [3,4,5]. The different protein variants from the same toxin, created under coevolution pressure, are generally called proteoforms – herein called toxiforms [6, 7]. Proteoform is a relatively new term, established by the top-down community, that complies all the different molecular forms in which the protein product of a single gene can be found, enclosing all isoforms, single-nucleotide polymorphism (SNP), genetic variation, alternative splicing of mRNA, and post-translational modifications (PTMs) [8]. In the universe of a venom sample, it is possible to estimate the existence of a great variety of toxiforms that can shift dynamically in time, under internal or external stimuli, or during toxin maturation processes.
Different proteomics approaches reveal the protein content of any venom. The most common and the gold standard method used nowadays is bottom-up proteomics (BUP) [9]. In all BUP strategies, proteins are digested in smaller peptides by enzymatic or chemical reactions and submitted to LC-MS/MS. The intact mass and fragmentation patterns are used to identify the peptides present in the sample according to a protein sequence database. From the identified peptides, it is possible to infer the toxin groups present in the venom or the occurrence of specific toxins through unique peptides. Peptide-centric based proteomics has been applied in toxinology studies since the time of 2D gel based proteomics to the latest cutting-edge techniques of shotgun proteomics [10, 11].
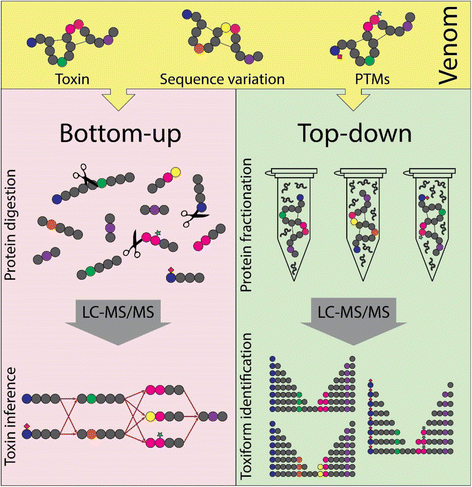
Focused on peptides, sometimes it is difficult for BUP strategies to infer the proteins present in a sample as well as to provide a biological interpretation of the data, especially when performing analysis of venom or toxic secretions. In these cases, the same peptide is often present in multiple different toxiforms. Such shared peptides, in most cases, lead to ambiguities in determining the identity of toxins (Fig. 1, left panel). This situation, called “protein inference problem”, clearly obscures the determination of the total number of toxiforms present in a venom [12].
Graphical representation of generic bottom-up (left panel) and top-down (right panel) venomics experiments. On the left panel, venom proteins are reduced, alkylated, enzymatically digested, and submitted to LC-MS/MS for peptide identification; toxins are inferred resulting in more protein possibilities than the original number of toxiforms (inference problem). On the right panel, venom is pre-fractionated before LC-MS/MS resulting in the identification and characterization of all toxiforms present in the initial sample
On the other hand, top-down proteomics (TDP), a method capable of measuring intact protein masses (without enzymatic digestion) and their fragment ions by MS, can provide the toxin information not reached with BUP techniques, as the identification and quantification of toxiforms and toxin complexes (Fig. 1, right panel) [13]. These approaches may help scientists to answer old questions on toxinology such as: “How many toxin variants – toxiforms – are present in a venom?”, “What is the degree of individual venom variance?”, or “What are the structural changes that take place during toxin maturation process?”. Therefore, TDP rises as the more informative technique to investigate venom proteome and toxiforms diversity. TDP methodologies are in development since the advent of soft ionization methods for MS in the late 1980s and can be applied to both denaturing and native TDP to determine venom proteomes [14,15,16,17].
Denaturing top-down proteomics
With more than 20 years of constant development and improvement, denaturing top-down proteomics (dTDP) is the most disseminated subarea of TDP. In this approach, at least once, a non-native condition – e.g. a denaturing substance (organic solvents, reducing agents, strong detergents, non-physiological pH, and others) – is used and/or a physical method (heat, pressure, etc.) that disrupts protein interactions and quaternary conformations. Usually, proteins are extracted in buffers containing strong detergents, chaotropic substances, and/or reducing agents before pre-fractionation using a denaturing method. Additionally, protein precipitation steps are also required to make the sample compatible with the next analyses. Then, fractions are submitted to LC-MS/MS, in which separation is performed by reversed-phase chromatography using organic solvents at low pH, making possible the identification of proteoforms and complex subunits present in the sample [16, 18].
dTDP has expanded fast in the last years and is reaching maturity for the analysis of proteins up to ~30 kDa, being capable to routinely perform qualitative and quantitative high-throughput analyses of intricate biological matrices in different proteomics laboratories worldwide [14, 19,20,21]. This achievement was possible due to recent advances in three important areas: protein fractionation, mass spectrometry, and data analysis.
Protein fractionation
Proteome dynamic range is generally vast, especially in venom samples in which it can reach up to four orders of magnitude [22]. Concomitantly, toxiforms expressed by multigene toxin families generally have similar molecular masses, making the venom a complex mixture of proteins. MS acquisition data cannot handle such diversity making necessary to pre-fractionate the venom prior to analysis [13]. However, intact proteins have the tendency to be less soluble than peptides and they have the inclination to stick to the stationary phase during chromatography, which is one of the major challenges for TDP [14].
Various techniques for protein fractionation have been used to separate intact proteoforms before MS; some of these methods are well known in biochemistry like reversed-phase liquid chromatography (RPLC), capillary isoelectric focusing (CIEF), size-exclusion chromatography (SEC), and capillary zone electrophoresis (CZE) [23,24,25,26,27,28]. Nevertheless, the most disseminated separation techniques in dTDP are solution isoelectric focusing (sIEF), and gel-eluted liquid fraction entrapment electrophoresis (GELFrEE), that can be used separately or combined, providing multidimensional fractionation before LC-MS/MS [29,30,31]. Recently, hydrophobic interaction chromatography (HIC) was coupled to MS and used as an alternative high-resolution separation to RPLC-MS [32].
Mass spectrometry
MS instruments used for TDP need to have high-resolving power, typically >50,000, to determine intact proteoform masses (MS1) and, especially for fragmentation spectrum (MS2), to correctly discriminate fragment ions that will be used for precise protein identification. Besides resolution, high mass accuracy, high sensitivity, and high speed are also important. Hybrid instruments that have time of flight (ToF), Fourier transform ion cyclotron resonance (FT-ICR), or FT-orbitraps as main mass analyzers achieve these aims; the last two are the most used in dTDP [33,34,35,36].
Orbitrap instruments are becoming the workhorses in dTDP because they are more cost effective and present promising hybrid architecture coupled to distinct fragmentation methods. Collision induced dissociation (CID) and high-energy collisional dissociation (HCD) are the classical fragmentation methods used in TDP studies [18]. However, electron transfer dissociation (ETD) [37], ultra violet photodissociation (UVPD) and the combination of more than one fragmentation type, e.g. EThcD and ETciD, are becoming more popular since they increase protein coverage and are available in new commercial instruments [38,39,40,41].
However, in spite of all these new capabilities, old problems limit the wide applicability of dTDP. Under denaturing conditions and using electrospray ionization (ESI), intact proteoforms show charge-state polydispersity (wider charge state envelopes). Additionally, the average number of protein charge states increases with the length of polypeptide chain causing ion signals split into several channels reducing the signal-to-noise ratio. In parallel, chemical noise from solvents and other substances used in sample handling, incomplete ion desolvation during ionization process, and presence of multiple PTMs in the same protein can frustrate the detection of proteoforms >30 kDa [42].
Venom – as a very complex mixture of proteins, mostly under 30 kDa depending on the venom source – requires high speed, high resolution and sensitivity to distinguish similar toxiforms. Moreover, only with high coverage of toxin fragmentation, it is possible to identify SNPs and PTMs of toxiforms not present in databases. Figure 2 illustrates the high fragmentation coverage obtained with TDP experiments of five toxiforms of acidic phospholipase A2 2 (Q9DF33) and two toxiforms of weak toxin DE-1 (P01412), both from the veom of Ophiophagus hannah that allowed correct proteoform identification and characterization [13].
Fragmentation maps of acidic phospholipase A2 2 (Q9DF33) and weak toxin DE-1 (P01412) toxiforms from Ophiophagus hannah venom. Gray squares represent amino acid alterations from the deposited sequence in UniProt database and the orange square represents a pyroglutamic acid. Data from Melani et al. [13] used to create this image are freely available at ProteomeExchange identifier PXD003403. Fragmentation maps, scoring and residue coverage were obtained using the software ProSight Lite
Data processing
With the increase in data collection, it is necessary to use software platforms to perform fast and confident automated processing of high resolution MS1 and MS2 data. The complex data obtained need to be deconvoluted to simplify the protein identification process and the softwares TRASH and MS-Deconv are commonly used [43, 44]. ProSight PTM was the first tool developed and further improved for a complete automated search using robust scores and statistics parameters to enable the identification and characterization of proteins, including coding polymorphisms, PTMs and proteoforms [45, 46]. Other search engines were also created as MS-Align+, MASH Suite, pTop, and TopPIC, as well as a new score for proteoform specificity, e.g., C-score [47,48,49,50,51].
ProSight PC (Thermo Scientific) is one of the most used tool that performs three distinct types of search: (1) absolute mass, similar to a PSM search with large tolerance window used for identification of proteoforms with PTMs; (2) biomarker search against all possible protein fragments within the database, similar to a BUP “non-enzymatic” search and ideal to identify protein cleavage; and (3) sequence tag search performing identification of proteins based on de novo sequencing from the fragmentation data, which is indicated for identification of proteins not included in a database [45]. In all search types p-score is calculated for each proteoform identification, representing the probability that a random sequence could account for the matching ions [52].
Software improvements made feasible high-throughput automated identification and characterization of several thousand proteoforms with high-confidence [20, 21, 53]. In TDP experiments, protein characterization and annotation must be based on MS/MS data supported by reliable scores and statistical analysis, including acceptable false discovery rate (FDR) values, as established for BUP analyses. Because of the high abundance of toxiforms, even for toxins from a single venom, it may be difficult to identify gene products based only in the intact mass and probable disulfide bonds. To identify undescribed toxiforms, MS2 fragmentation pattern and high sequence coverage, including the modified regions, are always required.
Denaturing top-down venomics
Verano-Braga et al., in 2013 [54], coined the term “top-down venomics” and used LC-MS/MS to identify, using de novo sequencing, peptides <10 kDa from the venom of Tityus serrulatus (Brazilian yellow scorpion). They obtained 1449 sequence tags of at least five amino acids from 73 proteins in total, by TDP approach, unraveling the role played by proteolysis in the molecular diversity of scorpion toxins [54]. In the same year, target top-down MALDI-ToF MS was used to sequence the toxin apamin isolated from the venom of Apis dorsata bee [55].
A first experimental attempt to apply dTDP to the study of snake venoms was made by Petras et al. [56] analyzing Ophiophagus hannah (king cobra) venom. A total of 15 intact toxins were manually identified by coupling LC-MS/MS analysis, intact mass values of reduced and non-reduced proteins, and BUP. Similar workflow based in locus specificity was applied to study the venom of Vipera anatolica (Anatolian meadow viper), Dendroaspis angusticeps (East African green mamba) and D. polylepis (black mamba) [57, 58].
The first high-throughput proteoform-centric dTDP study totally based in automated MS2 identification was performed on the venom of O. hannah by Melani et al. [13]. They applied different pre-fractionation techniques to identify 184 toxiforms from 131 proteins belonging to 14 toxin families. The data helped to clarify the view of sequence variation in three finger toxins, transit pro-peptide cleavage sites of ohanin and PTMs of venom toxins [13].
A key distinction between locus-centric versus toxiform-centric studies is that while the first simply seeks to identify a specific protein product present within the sample, proteoform analysis attempts to locate all sources of molecular variation amongst related toxiforms. Furthermore, unique peptides identified in BUP strategies are enough to assign protein locus, being not necessary TDP studies, which are more expensive and demanding to carry out.
Recently, Sanz-Medel’s group [59] published a promising workflow combining RPLC to inductively coupled plasma MS (ICP-MS) and denaturing MS for absolute quantitation and mass assignment of intact proteins. ICP-MS is a precise, accurate, and robust technique used in analytical chemistry to measure absolute isotope abundance of heteroatoms. Thus, isotope dilution analysis is performed adding 34S after protein fractionation and S content of proteins can be absolute measured. In parallel, mass profiling along the chromatographic separation is acquired by other MS, an ESI-Q-ToF, to provide protein molecular weight [59, 60].
When this method was applied to the venom of Naja mossambica (Mozambique spitting cobra) it was possible to quantify 27 intact masses of toxins [59]. However, the quantification is based on the premise of one protein/toxiform per chromatographic peak, which is not true for all the chromatographic fractions, as demonstrated in the SDS-PAGE of other study performed with the same venom [61]. Even more apprehensive is the fact that almost all “snake venomics” publications present SDS-PAGE figures of eluting RPLC fractions containing more than one toxin and/or toxiforms [62,63,64]. Eventually, peaks with more than one protein may produce toxin overestimation and errors in the protein concentration profile.
Venoms from snakes, scorpions, sea anemones, spiders, conus snails, bees, wasps, and other sources are rich in toxins with less than 30 kDa suitable to dTDP. Having in mind the large application of denaturing top-down venomics in the future and the number of identifications and characterizations of new toxiforms, it will be necessary to create new nomenclature rules and a repository site for the toxinology community. The Consortium for Top-down Proteomics (http://www.topdownproteomics.org/) has already made available a free repository where a venom database of O. hannah toxiforms is deposited [13].
Native top-down proteomics
While dTDP represent a current established proteomics technique, native top-down proteomics (nTDP), is a growing field [15, 17, 65]. Denaturing fractionation and ESI-MS are gentle enough to preserve covalent bonds and many covalent PTMs. However, the potentially biologically relevant non-covalent protein-protein and protein-ligand interactions are mostly destroyed. Quaternary states are conserved in nTDP using native protein extraction protocols, non-denaturing separation methods (without the use of denaturing chemical and physical agents), and native mass spectrometry. Consequently, nTDP can access, generally in single measurements, larger protein mass (> 50 kDa), subunit stoichiometry, binding associates, protein complex topology, labile PTMs, protein dynamics, and even binding affinities [66, 67].
Native MS analyses of protein complexes have been reported since the early 1990s using purified standard proteins and demonstrating that noncovalent interactions could be preserved in the gas phase when spraying aqueous solution at physiological pH [68,69,70]. Native MS offers the additional benefit of a lower distribution of charge states increasing signal-to-noise ratio because of lower number of channels to split ion intensity [16]. Early studies were carried out in triple quadrupole mass analyzers, followed by quadrupole time-of-flight (Q-ToF) mass analyzers. Recently, a modified orbitrap mass analyzer that allows the transmission of ions in the high m/z range was used in native MS as a more sensitive and higher resolution alternative [67].
Subunit ejection in the gas phase from homodimer complexes and the origin of asymmetric charge partitioning was only achieved and postulated in the beginning of the 2000s [71]. Late advances made possible, in benchtop quadrupole orbitraps, the complete characterization of protein complexes from their intact masses (MS1), subunit masses (MS2), and subunit fragmentation (MS3) opening a new possibility in nTDP field [72].
Applying the complete complex characterization method, Skinner and colleagues [73, 74] developed a native separation mode based in GELFrEE fractionation system, called native GELFrEE, that can fractionate complexes from endogenous systems prior to MS allowing to use nTDP in “discovery mode”. Following the same idea Muneeruddin et al. [75] coupled ion exchange chromatography on-line with native MS, potentially increasing the analysis throughput of unknown intact protein conjugates.
Together with method advancements in native fractionation and MS data acquisition there is the necessity for new bioinformatics tools for protein complex identification and characterization. A computational database search strategy was created by Neil Kelleher’s group [76], using an algorithm that considers intact, subunit and fragmentation masses, obtained by nTDP analysis, for precise identification and scoring of multi-proteoform complexes (MPC). With many analytical gains and ease access to biologically relevant proteoform interactions and masses, nTDP has the potential to change toxinology studies.
Native top-down venomics
Native fractionation and techniques to determinate protein-protein interactions are being applied to venom studies since classical works with crotoxin to recent studies that coupled SEC and denaturing MS [77, 78]. nTDP can be used in venom samples to identify large proteins and characterize macromolecular interactions among toxins by identification of complexes, their subunits, and PTMs.
Native top-down venomics was conceptualized and first applied to interrogate the venom of O. hannah [13]. Native GELFrEE fractionation and native MS analysis were combined to identify and characterize the glycosylated multichain toxin cobra venom factor (146 kDa), two clusters of glycosylated multiproteoform dimer of L-amino acid oxidase (126 and 130 kDa), a cysteine rich secretory protein homodimer (50 kDa), a phospholipase homodimer (26 kDa), and a metalloproteinase (49 kDa) [13].
With many toxins executing their functions as members of protein assemblies, observing biological organization and control at this hierarchical level will provide a more sophisticated view of the molecular composition of large toxiforms and protein-protein/protein-ligand interactions from venom multitoxiforms complexes.
Conclusions and perspectives
Top-down venomics is feasible and being applied in the last years to different venom sources even with some important bottlenecks in the areas of protein fractionation, mass spectrometry and software for data analysis. Future technical advances will make TDP more user-friendly, automated, and cheaper, helping to disseminate the technique throughout the scientific community.
Proteoform-centric dTDP is used in venomics studies and will be undoubtedly widely adopted in the toxinology field in the near future to help to answer new and old questions about venom variation, toxiforms and toxin processing/maturation. On the other hand, nTDP is more challenging to perform, demands top-end/customized mass spectrometers, and high-specialized trained personnel to perform experiments. However, it represents the future of top-down venomics because it provides information about large toxins, PTMs, and on protein interactions to unravel the MPCs world.
A precise molecular inventory of venom toxins obtained by TDP based in MS/MS techniques will expand our knowledge of the natural diversity of venom toxiforms. This will probably improve the quality and potency of antivenoms, uncover new molecular tools and new potential drugs, as well as provide initial steps needed to understand biological mechanisms the final goal of modern toxinology.
Abbreviations
- 2D:
-
Two dimensional
- BUP:
-
Bottom-up proteomics
- CID:
-
Collision induced dissociation
- CIEF:
-
Capillary isoelectric focusing
- CZE:
-
Capillary zone electrophoresis
- dTDP:
-
Denaturing top-down proteomics
- ESI:
-
Electrospray ionization
- ETciD:
-
Electron-transfer and collision induced dissociation
- ETD:
-
Electron transfer dissociation
- EThcD:
-
Electron-transfer and higher-energy collision dissociation
- FDR:
-
False discovery rate
- FT-ICR:
-
Fourier transform ion cyclotron resonance
- GELFrEE:
-
Gel-eluted liquid fraction entrapment electrophoresis
- HCD:
-
Higher-energy collisional dissociation
- HIC:
-
Hydrophobic interaction chromatography
- ICP-QQQ:
-
Inductively coupled plasma and triple quadrupole mass analyzer
- LC-MS/MS:
-
Liquid chromatography online with tandem mass spectrometry
- MALDI:
-
Matrix-assisted laser desorption/ionization
- MPC:
-
Multiproteoform complex
- mRNA:
-
Messenger ribonucleic acid
- MS:
-
Mass spectrometry
- MS/MS:
-
Tandem mass spectrometry
- nTDP:
-
Native top-down proteomics
- PTM:
-
Post-translational modification
- Q-ToF:
-
Quadrupole and time of flight .mass analyzer
- RPLC:
-
Reversed-phase liquid chromatography
- SDS-PAGE:
-
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SEC:
-
Size-exclusion chromatography
- sIEF:
-
Solution isoelectric focusing
- SNP:
-
Single-nucleotide polymorphism
- TDP:
-
Top-down proteomics
- ToF:
-
Time of flight
- UVPD:
-
Ultra violet photodissociation
References
Kardong KV. Snake toxins and venoms: an evolutionary perspective. Herpetologica. 1996;52(1):36–46.
de Lima ME, Pimenta AMC, Martin-Eauclaire MF, Zingali RB, Rochat H. Animal toxins: state of the art. Perspectives in health and biotechnology. 1st ed. UFMG: Belo Horizonte; 2009.
Kordis D, Gubensek F. Adaptive evolution of animal toxin multigene families. Gene. 2000;261(1):43–52.
Fox JW, Serrano SMT. Exploring snake venom proteomes: multifaceted analyses for complex toxin mixtures. Proteomics. 2008;8(4):909–20.
Georgieva D, Arni RK, Betzel C. Proteome analysis of snake venom toxins: pharmacological insights. Expert Rev Proteomics. 2008;5(6):787–97.
Holding ML, Biardi JE, Gibbs HL. Coevolution of venom function and venom resistance in a rattlesnake predator and its squirrel prey. Proc Biol Sci. 2016;283(1829):20152841.
Hanifin CT, Brodie ED Jr, Brodie ED. Phenotypic mismatches reveal escape from arms-race coevolution. PLoS Biol. 2008;6(3):e60.
Smith LM, Kelleher NL, Linial M, Goodlett D, Langridge-Smith P, Goo A. Y, et al. Proteoform: a single term describing protein complexity. Nat Methods. 2013;10(3):186–7.
Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR. Protein analysis by shotgun/bottom-up proteomics. Chem Rev. 2013;113(4):2343–94.
Melani RD, Goto-Silva L, Nogueira FCS, Junqueira M, Domont GB. Shotgun approaches for venom analysis. In: Gopalakrishnakone P, Calvete JJ, editors. Venom genomics proteomics. Netherlands: Springer; 2014. p. 1–12.
Lomonte B, Calvete JJ. Strategies in 'snake venomics' aiming at an integrative view of compositional, functional, and immunological characteristics of venoms. J Venom Anim Toxins incl Trop Dis. 2017;23:26. doi:10.1186/s40409-017-0117-8.
Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol Cell Proteomics. 2005;4(10):1419–40.
Melani RD, Skinner OS, Fornelli L, Domont GB, Compton PD, Kelleher NL. Mapping proteoforms and protein complexes from king cobra venom using both denaturing and native top-down proteomics. Mol Cell Proteomics. 2016;15(7):2423–34.
Toby TK, Fornelli L, Kelleher NL. Progress in top-down proteomics and the analysis of proteoforms. Annu Rev Anal Chem (Palo Alto, Calif). 2016;9(1):499–519.
Fornelli L, Toby TK, Schachner LF, Doubleday PF, Srzentić K, DeHart CJ, et al. Top-down proteomics: where we are, where we are going? J Proteome. 2017; doi:10.1016/j.jprot.2017.02.002.
Catherman AD, Skinner OS, Kelleher NL. Top down proteomics: facts and perspectives. Biochem Biophys Res Commun. 2014;445(4):683–93.
Compton PD, Kelleher NL. Spinning up mass spectrometry for whole protein complexes. Nat Methods. 2012;9(11):1065–6.
Patrie SM. Top-down mass spectrometry: proteomics to Proteoforms. In: Mirzaei H, Carrasco M, editors. Mod. Proteomics - sample prep anal Pract Appl: Springer international publishing; 2016. p. 171–200. doi:10.1007/978-3-319-41448-5 8.
Ntai I, Kim K, Fellers RT, Skinner OS, Smith AD, Early BP, et al. Applying label-free quantitation to top down proteomics. Anal Chem. 2014;86(10):4961–8.
Anderson JC, Wan Y, Kim YM, Pasa-Tolic L, Metz TO, Peck SC. Decreased abundance of type III secretion system-inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae. Proc Natl Acad Sci. 2014;111(18):6846–51.
Durbin KR, Fornelli L, Fellers RT, Doubleday PF, Narita M, Kelleher NL. Quantitation and identification of thousands of human proteoforms below 30 kDa. J Proteome Res. 2016;15(3):976–82.
Melani RD, Araujo GDT, Carvalho PC, Goto L, Nogueira FCS, Junqueira M, et al. Seeing beyond the tip of the iceberg: a deep analysis of the venome of the Brazilian rattlesnake, Crotalus durissus terrificus. EuPA Open Proteom. 2015;8:144–56.
Shen Y, Tolić N, Zhao R, Paša-Tolić L, Li L, Berger SJ, et al. High-throughput proteomics using high-efficiency multiple-capillary liquid chromatography with on-line high-performance ESI FTICR mass spectrometry. Anal Chem. 2001;73(13):3011–21.
Lee SW, Berger SJ, Martinović S, Paša-Tolić L, Anderson GA, Shen Y, et al. Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc Natl Acad Sci U S A. 2002;99(9):5942–7.
Martinović S, Berger SJ, Paša-Tolić L, Smith RD. Separation and detection of intact noncovalent protein complexes from mixtures by on-line capillary isoelectric focusing-mass spectrometry. Anal Chem. 2000;72(21):5356–60.
Simpson DC, Ahn S, Pasa-Tolic L, Bogdanov B, Mottaz HM, Vilkov AN, et al. Using size exclusion chromatography-RPLC and RPLC-CIEF as two-dimensional separation strategies for protein profiling. Eletrophoresis. 2006;27(13):2722–33.
Valaskovic GA, Kelleher NL, McLafferty FW. Attomole protein characterization by capillary electrophoresis-mass spectrometry. Science. 1996;273(5279):1199–202.
Li Y, Compton PD, Tran JC, Ntai I, Kelleher NL. Optimizing capillary electrophoresis for top-down proteomics of 30-80 kDa proteins. Proteomics. 2014;14(10):1158–64.
Tran JC, Doucette AA. Rapid and effective focusing in a carrier ampholyte solution isoelectric focusing system: a proteome prefractionation tool. J Proteome Res. 2008;7(4):1761–6.
Tran JC, Doucette AA. Gel-eluted liquid fraction entrapment electrophoresis: an electrophoretic method for broad molecular weight range proteome separation. Anal Chem. 2008;80(5):1568–73.
Tran JC, Zamdborg L, Ahlf DR, Lee JE, Catherman AD, Durbin KR, et al. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature. 2011;480(7376):254–8.
Chen B, Peng Y, Valeja SG, Xiu L, Alpert AJ, Ge Y. Online hydrophobic interaction chromatography-mass spectrometry for top-down proteomics. Anal Chem. 2016;88(3):1885–91.
Fornelli L, Parra J, Hartmer R, Stoermer C, Lubeck M, Tsybin YO. Top-down analysis of 30-80 kDa proteins by electron transfer dissociation time-of-flight mass spectrometry. Anal Bioanal Chem. 2013;405(26):8505–14.
Chamot-Rooke J, Mikaty G, Malosse C, Soyer M, Dumont A, Gault J, et al. Posttranslational modification of pili upon cell contact triggers N. meningitidis dissemination. Science. 2011;331(6018):778–82.
Bogdanov B, Smith RD. Proteomics by FTICR mass spectrometry: top down and bottom up. Mass Spectrom Rev. 2005;24(2):168–200.
Perry RH, Cooks RG, Noll RJ. Orbitrap mass spectrometry: instrumentation, ion motion and applications. Mass Spectrom Rev. 2008;27(6):661–99.
Bem Hamidane H, Chiappe D, Hartmer R, Vorobyev A, Moniatte M, Tsybin YO. Electron capture and transfer dissociation: peptide structure analysis at different ion internal energy levels. J Am Soc Mass Spectrom. 2009;20(4):567–75.
Cannon JR, Cammarata MB, Robotham SA, Cotham VC, Shaw JB, Fellers RT, et al. Ultraviolet photodissociation for characterization of whole proteins on a chromatographic time scale. Anal Chem. 2014;86(4):2185–92.
Brunner AM, Lössl P, Liu F, Huguet R, Mullen C, Yamashita M, et al. Benchmarking multiple fragmentation methods on an orbitrap fusion for top-down phospho-proteoform characterization. Anal Chem. 2015;87(8):4152–8.
Fornelli L, Damoc E, Thomas PM, Kelleher NL, Aizikov K, Denisov E, et al. Analysis of intact monoclonal antibody IgG1 by electron transfer dissociation orbitrap FTMS. Mol Cell Proteomics. 2012;11(12):1758–67.
Cleland TP, DeHart CJ, Fellers RT, VanNispen AJ, Greer JB, LeDuc RD, et al. High-throughput analysis of intact human proteins using UVPD and HCD on an orbitrap mass spectrometer. J Proteome Res. 2017;16(5):2072–9.
Compton PD, Zamdborg L, Thomas PM, Kelleher NL. On the scalability and requirements of whole protein mass spectrometry. Anal Chem. 2011;83(17):6868–74.
Horn DM, Zubarev RA, McLafferty FW. Automated reduction and interpretation of high resolution electrospray mass spectra of large molecules. J Am Soc Mass Spectrom. 2000;11(4):320–32.
Liu X, Inbar Y, Dorrestein PC, Wynne C, Edwards N, Souda P, et al. Deconvolution and database search of complex tandem mass spectra of intact proteins: a combinatorial approach. Mol Cell Proteomics. 2010;9(12):2772–82.
Zamdborg L, LeDuc RD, Glowacz KJ, Kim YB, Viswanathan V, Spaulding IT, et al. ProSight PTM 2.0: improved protein identification and characterization for top down mass spectrometry. Nucleic Acids Res. 2007;35:W701–6.
LeDuc RD, Taylor GK, Kim YB, Januszyk TE, Bynum LH, Sola JV, et al. ProSight PTM: an integrated environment for protein identification and characterization by top-down mass spectrometry. Nucleic Acids Res. 2004;32:W340–5.
Liu X, Sirotkin Y, Shen Y, Anderson G, Tsai YS, Ting YS, et al. Protein identification using top-down spectra. Mol Cell Proteomics. 2012;11(6):M111.008524.
Guner H, Close PL, Cai W, Zhang H, Peng Y, Gregorich ZR, et al. MASH suite: a user-friendly and versatile software interface for high-resolution mass spectrometry data interpretation and visualization. J Am Soc Mass Spectrom. 2014;25(3):464–70.
Sun RX, Luo L, Wu L, Wang RM, Zeng WF, Chi H, et al. pTop 1.0: a high-accuracy and high-efficiency search engine for intact protein identification. Anal Chem. 2016;88(6):3082–90.
Kou Q, Xun L, Liu X. TopPIC: a software tool for top-down mass spectrometry-based proteoform identification and characterization. Bioinformatics. 2016;32(22):3495–7.
LeDuc RD, Fellers RT, Early BP, Greer JB, Thomas PM, Kelleher NL. The C-score: a Bayesian framework to sharply improve proteoform scoring in high-throughput top down proteomics. J Proteome Res. 2014;13(7):3231–40.
Meng F, Cargile BJ, Miller LM, Forbes AJ, Johnson JR, Kelleher NL. Informatics and multiplexing of intact protein identification in bacteria and the archaea. Nat Biotechnol. 2001;19(10):952–7.
Vorontsov EA, Rensen E, Prangishvili D, Krupovic M, Chamot-Rooke J. Abundant lysine methylation and N-terminal acetylation in Sulfolobus islandicus revealed by bottom-up and top-down proteomics. Mol Cell Proteomics. 2016;15(11):3388–404.
Verano-Braga T, Dutra AAA, León IR, Melo-Braga MN, Roepstorff P, Pimenta AMC, et al. Moving pieces in a venomic puzzle: unveiling post-translationally modified toxins from Tityus serrulatus. J Proteome Res. 2013;12(7):3460–70.
Baracchi D, Mazza G, Michelucci E, Pieraccini G, Turillazzi S, Moneti G. Top-down sequencing of Apis dorsata apamin by MALDI-TOF MS and evidence of its inactivity against microorganisms. Toxicon. 2013;71:105–12.
Petras D, Heiss P, Süssmuth RD, Calvete JJ. Venom proteomics of Indonesian king cobra, Ophiophagus hannah: integrating top-down and bottom-up approaches. J Proteome Res. 2015;14(6):2539–56.
Göçmen B, Heiss P, Petras D, Nalbantsoy A, Süssmuth RD. Mass spectrometry guided venom profiling and bioactivity screening of the Anatolian Meadow Viper, Vipera anatolica. Toxicon. 2015;107(Pt B):163–74.
Petras D, Heiss P, Harrison RA, Süssmuth RD, Calvete JJ. Top-down venomics of the east African green mamba, Dendroaspis angusticeps, and the black mamba, Dendroaspis polylepis, highlight the complexity of their toxin arsenals. J Proteome. 2016;146(2):148–64.
Calderón-Celis F, Diez-Fernández S, Costa-Fernández JM, Encinar JR, Calvete JJ, Sanz-Medel A. Elemental mass spectrometry for absolute intact protein quantification without protein-specific standards: application to snake venomics. Anal Chem. 2016;88(19):9699–706.
Calvete JJ, Petras D, Calderón-Celis F, Lomonte B, Encinar JR, Sanz-Medel A. Protein-species quantitative venomics: looking through a crystal ball. J Venom Anim Toxins incl Trop Dis. 2017;23:27. doi:10.1186/s40409-017-0116-9.
Petras D, Sanz L, Segura A, Herrera M, Villalta M, Solano D, et al. Snake venomics of African spitting cobras: toxin composition and assessment of congeneric cross-reactivity of the pan-African EchiTAb-plus-ICP antivenom by antivenomics and neutralization approaches. J Proteome Res. 2011;10(3):1266–80.
Tashima AK, Sanz L, Camargo ACM, Serrano SMT, Calvete JJ. Snake venomics of the Brazilian pitvipers Bothrops cotiara and Bothrops fonsecai. Identification of taxonomy markers. J Proteome. 2008;71(4):473–85.
Boldrini-França J, Corrêa-Netto C, Silva MMS, Rodrigues RS, De La Torre P, Pérez A, et al. Snake venomics and antivenomics of Crotalus durissus subspecies from Brazil: assessment of geographic variation and its implication on snakebite management. J Proteome. 2010;73(9):1758–76.
Rodrigues RS, Boldrini-França J, Fonseca FPP, de la Torre P, Henrique-Silva F, Sanz L, et al. Combined snake venomics and venom gland transcriptomic analysis of Bothropoides pauloensis. J Proteome. 2012;75(9):2707–20.
Haverland NA, Skinner OS, Fellers RT, Tariq AA, Early BP, RD LD, et al. Defining gas-phase fragmentation propensities of intact proteins during native top-down mass spectrometry. J Am Soc Mass Spectrom. 2017;28:1203–15.
Lössl P, Waterbeemd M. Van de, heck AJ. The diverse and expanding role of mass spectrometry in structural and molecular biology. EMBO J. 2016;35(24):2634–57.
Leney AC, Heck AJR. Native mass spectrometry: what is in the name? J Am Soc Mass Spectrom. 2017;28(1):5–13.
Ganem B, Li YT, Henion JD. Detection of noncovalent receptor-ligand complexes by mass spectrometry. J Am Chem Soc. 1991;113(16):6294–6.
Katta V, Chait BT. Observation of the heme-globin complex in native myoglobin by electrospray-ionization mass spectrometry. J Am Chem Soc. 1991;113(22):8534–5.
Li YT, Hsieh YL, Henion JD, Senko MW, McLafferty FW, Ganem B. Mass spectrometric studies on noncovalent dimers of leucine zipper peptides. J Am Chem Soc. 1993;115(18):8409–13.
Jurchen JC, Williams ER. Origin of asymmetric charge partitioning in the dissociation of gas-phase protein homodimers. J Am Chem Soc. 2003;125(9):2817–26.
Belov ME, Damoc E, Denisov E, Compton PD, Horning S, Makarov AA, et al. From protein complexes to subunit backbone fragments: a multi-stage approach to native mass spectrometry. Anal Chem. 2013;85(23):11163–73.
Skinner OS, Do Vale LHF, Catherman AD, Havugimana PC, de Sousa MV, Compton PD, et al. Native GELFrEE: a new separation technique for biomolecular assemblies. Anal Chem. 2015;87(5):3032–8.
Melani RD, Seckler HS, Skinner OS, do Vale LHFD, Catherman AD, Havugimana PC, et al. CN-GELFrEE - clear native gel-eluted liquid fraction entrapment electrophoresis. J Vis Exp. 2016;29(108):e53597.
Muneeruddin K, Nazzaro M, Kaltashov IA. Characterization of intact protein conjugates and biopharmaceuticals using ion-exchange chromatography with online detection by native electrospray ionization mass spectrometry and top-down tandem mass spectrometry. Anal Chem. 2015;87(19):10138–45.
Skinner OS, Havugimana PC, Haverland NA, Fornelli L, Early BP, Greer JB, et al. An informatic framework for decoding protein complexes by top-down mass spectrometry. Nat Methods. 2016;13(3):237–40.
Fraenkel-Conrat H, Singer B. Fractionation and composition of crotoxin. Arch Biochem Biophys. 1956;60(1):64–73.
Fernández J, Vargas-Vargas N, Pla D, Sasa M, Rey-Suárez P, Sanz L, et al. Snake venomics of Micrurus alleni and Micrurus mosquitensis from the Caribbean region of Costa Rica reveals two divergent compositional patterns in new world elapids. Toxicon. 2015;107(Pt B):217–33.
Acknowledgements
The authors would like to thanks CAPES, CNPq, FARPEJ, and Petrobras.
Funding
RDM is National Council for Scientific and Technological Development (CNPq), Brazil, post-doctoral fellow under award number 150232/2016–0. FCSN is FAPERJ fellow, grant E-26/202.801/2015. GBD is fellow of the National Council for Scientific and Technological Development, CNPq, Brazil (Grant 306,316/2015–3).
Author information
Authors and Affiliations
Contributions
RDM drafted the manuscript and all authors contributed writing and revising the manuscript. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Melani, R.D., Nogueira, F.C.S. & Domont, G.B. It is time for top-down venomics. J Venom Anim Toxins Incl Trop Dis 23, 44 (2017). https://doi.org/10.1186/s40409-017-0135-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40409-017-0135-6